New Pfizer antiviral and ivermectin, a pharmacodynamic analysis
New Pfizer antiviral, PF-07321332, C₂₃H₃₂F₃N₅O₄
PF-07321332 is designed to block the activity of the SARS-CoV-2-3CL protease,
https://www.pfizer.com/news/press-rel…
So, what is a protease?
So what is a protease inhibitor?
And, what is 3CL?
Chymotrypsin-like protease (3CL main protease, or 3CL Mpro)
Identification of SARS-CoV‑2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening (3rd September 2020)
https://pubs.acs.org/doi/abs/10.1021/…
The activity of the anti-SARS-CoV-2 viral infection was confirmed in 7 of 23 compounds
Microscopic interactions between ivermectin and key human and viral proteins involved in SARS-CoV-2 infection
https://pubs.rsc.org/en/content/artic…
the strength and persistency of the interaction between IVE and the binding site of 3CLpro indicate that a partial inhibition of the catalytic activity could have place as the drug interacts with the main subdomains that define the enzyme binding pocket:
Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents
https://www.nature.com/articles/s4200…
as shown in Fig. 4, out of 13 OTDs only ivermectin completely blocked ( more than 80%) the 3CLpro activity at 50 µM concentration.
Development, validation, and approval of COVID-19 specific drugs takes years. Therefore, the idea of drug repositioning, also known as repurposing, is an important strategy to control the sudden outbreak of life-threatening infectious agents that spread rapidly.
Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: designing, docking and molecular dynamics simulation study
https://pubs.rsc.org/en/content/artic…
From the docking analysis, ivermectin showed the highest docking score with an average energy of −8.5 kcal mol−1 among all the compounds. Remdesivir showed the lowest binding energy and highest docking score of −9.9 kcal mol−1
https://bnf.nice.org.uk/medicinal-for…
Ritonavir, C37H48N6O5S2
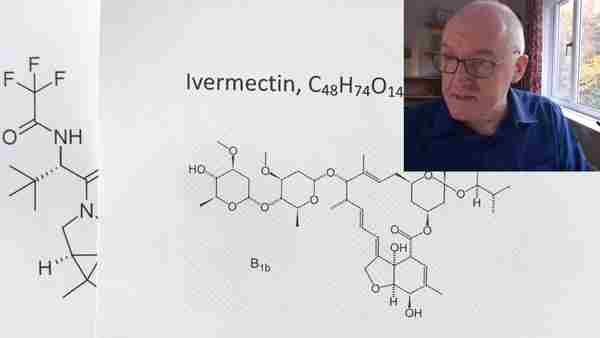
Ivermectin, C48H74O14
Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach
https://www.ncbi.nlm.nih.gov/pmc/arti…
We have documented an intense binding of both ivermectin B1a and B1b isomer to the main protease with subsequent energy (ETot-) values of -384.56 and -408.6.
PF-07321332 is designed to block the activity of the SARS-CoV-2-3CL protease,
https://www.pfizer.com/news/press-rel…
Risk of virus developing resistance to PF-07321332
Molecular Docking Reveals Ivermectin and Remdesivir as Potential Repurposed Drugs Against SARS-CoV-2
https://www.frontiersin.org/articles/…
With SARS-CoV-2 S Spike protein
Ivermectin showed high binding affinity to the viral S protein as well as the human cell surface receptors ACE-2 and TMPRSS2.
In agreement to our findings, ivermectin was found to be docked between the viral spike and the ACE2 receptor
Binding Interactions of Selected Drugs With Human TMPRSS2 Protein (ACE2 protein)
The docking results revealed that ivermectin showed the highest binding affinity to the active site of the protein (MolDock score −174.971) and protein–ligand interactions
Binding Interactions of Selected Drugs With Human ACE-2 Protein
that ivermectin showed the highest binding affinity to the active site of the protein (MolDock score −159.754) and protein–ligand interactions
With SARS-CoV-2 S Glycoprotein
Ivermectin showed the highest binding affinity to the predicted active site of the protein
With SARS-CoV-2 Nsp14 Protein
ivermectin showed the highest binding affinity (MolDock score −212.265) and protein–ligand interactions
Binding Interactions of Selected Drugs With SARS-CoV-2 PLpro
Ivermectin showed the highest binding affinity to the predicted active site of the protein (MolDock score −180.765) and protein–ligand interactions
A brief message to world leaders
Come on ya all