Summary
The mRNA-based BNT162b2 vaccine from Pfizer/BioNTech was the first registered COVID-19 vaccine and has been shown to be up to 95% effective in preventing SARS-CoV-2 infections. Little is known about the broad effects of the new class of mRNA vaccines, especially whether they have combined effects on innate and adaptive immune responses. Here we confirmed that BNT162b2 vaccination of healthy individuals induced effective humoral and cellular immunity against several SARS-CoV-2 variants. Interestingly, however, the BNT162b2 vaccine also modulated the production of inflammatory cytokines by innate immune cells upon stimulation with both specific (SARS-CoV-2) and non-specific (viral, fungal and bacterial) stimuli. The response of innate immune cells to TLR4 and TLR7/8 ligands was lower after BNT162b2 vaccination, while fungi-induced cytokine responses were stronger. In conclusion, the mRNA BNT162b2 vaccine induces complex functional reprogramming of innate immune responses, which should be considered in the development and use of this new class of vaccines.
Main text
Coronavirus disease 2019 (COVID-19) is a new respiratory tract infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which spread worldwide since the end of 2019 causing a global pandemic. The COVID-19 pandemic represents the most important healthcare crisis humanity has encountered since World War II, combined with a devastating societal and economic impact. Confronted with this critical situation, a major effort to develop vaccines against COVID-19 has been initiated in many countries around the world.
To date, 13 vaccines have been approved for use in humans (“COVID-19 vaccine tracker | RAPS,” n.d.). The scale of the pandemic has led to accelerated development of vaccines based on new technologies, such as mRNA- and viral vector-based vaccines (van Riel & de Wit, 2020). One of the most widely used anti-COVID-19 vaccines in the world was developed by a collaboration between BioNTech and Pfizer (BNT162b2). This vaccine is based on a lipid nanoparticle–formulated, nucleoside-modified mRNA that encodes a prefusion stabilized form of the spike (S)-protein derived from the SARS-CoV-2 strain isolated early on in Wuhan, China (Walsh et al., 2020). Several phase-3 trials have demonstrated that BNT162b2 elicits broad humoral and cellular responses, providing protection against COVID-19 (Sahin et al., 2020; Walsh et al., 2020).
While global vaccination campaigns against the SARS-CoV-2 infection are rolled out, major challenges remain, especially the spread of novel virus variants (Madhi et al., 2021). One of the most prominent mutations during the pandemic has been the spike D614G substitution to the Wuhan Hu-1 original strain (Korber et al., 2020). With the steadily increasing prevalence of infections, SARS-CoV-2 variants emerged with multiple spike mutations and were first detected in the United Kingdom (B.1.1.7 lineage), South Africa (B.1.351 lineage), and Brazil (P.1 lineage). These variants are of significant concern because of their potential effects on disease severity, viral transmissibility, reinfection rates, and vaccine effectiveness (Abdool Karim & de Oliveira, 2021).
The capacity of BNT162b2 to induce effective humoral and cellular immunity against the new SARS-CoV-2 variants is only now beginning to be understood. Whereas neutralization of B.1.1.7 was either similar or just slightly reduced as compared to the standard strain (Muik et al., 2021; Wang, Nair, et al., 2021), neutralizing titers of B.1.351 were markedly diminished (Liu et al., 2021; Planas et al., 2021; Wang, Nair, et al., 2021) after vaccination of healthy volunteers with BNT162b2. In contrast, cellular immunity against the virus variants seems to be less affected (Lilleri et al., 2021; Skelly et al., 2021). In addition, an unexplored area is whether vaccination with BNT162b2 also leads to long-term effects on innate immune responses: this could be very relevant in COVID-19, in which dysregulated inflammation plays an important role in the pathogenesis and severity of the disease (Tahaghoghi-Hajghorbani et al., 2020). The long-term modulation of innate immune responses has been an area of increased interest in the last years: multiple studies have shown that long-term innate immune responses can be either increased (trained immunity) or down-regulated (innate immune tolerance) after certain vaccines or infections (Netea et al., 2020).
In this study, we assessed the effect of the BNT162b2 mRNA COVID-19 vaccine on both the innate and adaptive (humoral and cellular) immune responses. We first examined the concentration of RBD- and S-binding antibody isotype concentrations before vaccination (baseline; t1), 3 weeks after the first dose of 30 μg of BNT162b2 (t2), and two weeks after the second dose (t3) (Figure S1A). We calculated fold-changes by comparing concentrations from both t2 and t3 to baseline. BNT162b2 vaccination elicited high anti-S protein and anti-RBD antibody concentrations already after the first vaccination, and even stronger responses after the second dose of the vaccine. As expected, IgG responses were the most pronounced, with RBD-specific median fold changes at t2 and t3 of 56-fold and 1839-fold, and S-specific fold changes of 208-fold and 1100-fold, respectively. The lowest observed fold change increase to pre-vaccination levels of IgG targeting RBD was 14-fold at t2, and 21-fold at t3, respectively. For S-specific IgG, the fold changes were at least 32-fold at t2 and 339-fold at t3. Regarding IgA concentrations, a single dose of the vaccine elicited a 7-fold increase in the RBD-specific concentration and a 35-fold increase in the S-specific concentration. The second dose enhanced the antibody concentrations elicited by the first vaccination by 24-fold and 52-fold, for RBD and S, respectively. Compared to IgG and IgA, the increase in IgM concentrations was considerably lower. RBD-specific concentration only doubled after the first dose, and it did not further increase after the second dose of the vaccine. In contrast, S-specific fold changes were 11-fold at t2 and 20-fold at t3 (Figure S1A). These results confirm and extend recent observations reporting strong induction of humoral responses by BNT162b2 vaccination (Sahin et al., 2020).
To investigate the neutralizing capacity of the serum against SARS-CoV-2 variants, we performed 50% plaque reduction neutralization testing (PRNT50) using sera collected two weeks after the second vaccine administration. All the serum samples neutralized the D614G strain and the B.1.1.7 variant with titers of at least 1:80. However, six subjects (37,5%) had titers lower than 1:80 against the B.1.351 variant. Geometric mean neutralizing titers against the D614G strain, B.1.1.7 and B.1.351 were 381, 397, and 70, respectively (Figure S1B, p<0.001). Similar to our investigation, several studies reported 6 to 14-fold decreased neutralizing activity of post-vaccine sera against the B.1.351 variant, and only slightly reduced activity against B.1.1.7, when compared to the standard strain (Planas et al., 2021; Shen et al., 2021; Wang, Liu, et al., 2021). These data support the evidence that B.1.351, and possibly other variants, may be able to escape vaccine-induced humoral immunity to a certain extent (Kustin et al., 2021). Furthermore, the PRNT titer and the antibody concentrations of IgG after the second dose were strongly correlated (Figure S1C). The correlation was stronger for B.1.1.7 and B.1.351 than for the standard strain, both for anti-RBD and anti-S.
BNT162b2 vaccination has been reported to activate virus-specific CD4+ and CD8+ T cells, and upregulate the production of immune-modulatory cytokines such as IFN-γ (Sahin et al., 2020). Hence, we assessed IFN-γ secretion from peripheral blood mononuclear cells (PBMCs) before and after BNT162b2 vaccination in response to heat-inactivated SARS-CoV-2 strains (Figs 2A-2D). While vaccination with BNT162b2 generally seems to moderately increase specific IFN-γ production after the second dose of the vaccine, this reached statistical significance only upon stimulation with B.1.351 variant (Figure S2A-2C). The same tendency has also been observed by Tarke et al. who used synthetic SARS-CoV-2 variants proteins to induce elevated IFN-γ responses against B.1.351 (Tarke et al., 2021). IFN-γ production was higher by at least 50% in 37.5% of the subjects upon stimulation with the standard SARS-CoV-2 strain, in 50% of the subjects upon stimulation with the B.1.1.7 variant and the B.1.351 variant, but only in 18,75% of the subjects upon stimulation with the Bavarian variant (Figure S1D). These findings argue that BNT162b2 vaccination induces better humoral than cellular immune responses. Weak T-cell responses have previously been reported in vaccinees that have received just a single dose of BNT162b2 (Prendecki et al., 2021; Stankov, Cossmann, Bonifacius, Dopfer-jablonka, & Morillas, 2021). Intriguingly, the best cellular responses after vaccination were against the B.1.351 variant: the fact that the neutralizing antibody responses against this variant were relatively poor, that may raise the possibility that the protective BNT162b2 vaccine effects against this variant may be mainly reliant on cellular, rather than humoral, responses. No significant differences between the individual variants were observed. The absolute concentrations of the cytokines after stimulations can be found in Supplementary Table 2.
Interestingly, we observed important heterologous effects of BNT162b2 vaccination on IFN-γ production induced by other stimuli as well (Figures S2E, 2F). BNT162b2 vaccination decreased IFN-γ production upon stimulation with the TLR7/8 agonist R848 (Figure S2F). In contrast, the IFN-γ production induced by inactivated influenza virus tended to be higher two weeks after the second BNT162b2 vaccination, though the differences did not reach statistical significance. We did not find any significant correlation between cellular responses and IgG antibody titers.
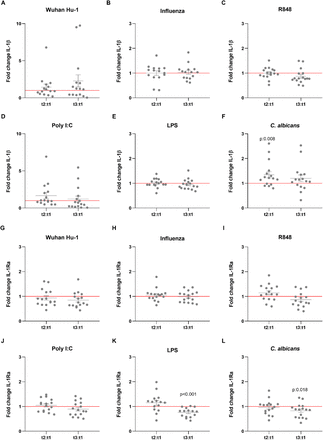
Besides their effects on specific (adaptive) immune memory, certain vaccines such as Bacillus Calmette-Guérin (BCG) and the measles, mumps, and rubella (MMR) vaccine also induce long-term functional reprogramming of cells of the innate immune system. (Netea et al., 2020). This biological process is also termed trained immunity when it involves increased responsiveness, or innate immune tolerance when it is characterized by decreased cytokine production (Ifrim et al., 2014). Although these effects have been proven mainly for live attenuated vaccines, we sought to investigate whether the BNT162b2 vaccine might also induce effects on innate immune responses against different viral, bacterial and fungal stimuli. One of the trademarks of trained immunity is an elevated production of inflammatory cytokines following a secondary insult (Quintin et al., 2012). Surprisingly, the production of the monocyte-derived cytokines TNF-α, IL-1β and IL-1Ra tended to be lower after stimulation of PBMCs from vaccinated individuals with either the standard SARS-CoV-2 strain or heterologous Toll-like receptor ligands (Figures 1 and 2). TNF-α production (Figure 1B-1G) following stimulation with the TLR7/8 agonist R848 of peripheral blood mononuclear cells from volunteers was significantly decreased after the second vaccination (Figure 1C). The same trend was observed after stimulation with the TLR3 agonist poly I:C (Figure 1D), although the difference did not reach statistical significance. In contrast, the responses to the fungal pathogen Candida albicans were higher after the first dose of the vaccine (Figure 1G). The impact of the vaccination on IL-1β production was more limited (Figure 2A-2F), though the response to C. albicans was significantly increased (Figure 2F). The production of the anti-inflammatory cytokine IL-1Ra (Figure 2G-2L) was reduced in response to bacterial lipopolysaccharide (LPS) and C. albicans after the second vaccination (Figure 2K, 2L), which is another argument for a shift towards stronger inflammatory responses to fungal stimuli after vaccination. IL-6 responses were similarly decreased, though less pronounced (data not shown).
(A) Description of the study: vaccination and blood collection days. (B-G) Fold change values of TNF-α production are calculated individually for each subject by division of t2:t1 and t3:t1. Data are presented as fold changes ± SEM (n=15-16) and analysed by Wilcoxon’s matched-pairs signed-rank test comparing each ratio to t1=1 (red line). (H-I) IFN-α production (pg/ml) at t1, t2 and t3. Data are presented as cytokine concentration ± SEM (n=15-16) and analysed by Wilcoxon’s matched-pairs signed-rank test.
IL-1β (A-F) and IL-1Ra (G-L) Fold change values are calculated individually for each subject by division of t2:t1 and t3:t1. Data are presented as fold changes ± SEM (n=15-16) and analysed by Wilcoxon’s matched-pairs signed-rank test comparing each ratio to t1=1 (red line).
The induction of tolerance towards stimulation with TLR7/8 (R848) or TLR4 (LPS) ligands by BNT162b2 vaccination may indicate a more balanced inflammatory reaction during infection with SARS-CoV-2, and one could speculate whether such effect may be thus useful to regulate the potential over-inflammation in COVID-19, one of the main causes of death (Tang et al., 2020). On the other hand, inhibition of innate immune responses may diminish anti-viral responses. Type I interferons also play a central role in the pathogenesis and response against viral infections, including COVID-19 (Hadjadj et al., 2020). With this in mind, we also assessed the production of IFN-α by immune cells of the volunteers after vaccination. Although the concentrations of IFN-α were below the detection limit of the assay for most of the stimuli, we observed a significant reduction in the production if IFN-α secreted after stimulation with poly I:C and R848 after the administration of the second dose of the vaccine (Figure 1H, 1I). This may hamper the initial innate immune response against the virus, as defects in TLR7 have been shown to result in and increased susceptibility to COVID-19 in young males (Van Der Made et al., 2020). These results collectively demonstrate that the effects of the BNT162b2 vaccine go beyond the adaptive immune system and can also modulate innate immune responses.
The effect of the BNT162b2 vaccination on innate immune responses may also indicate a potential to interfere with the responses to other vaccinations, as known for other vaccines to be as ‘vaccine interference’ (Lum et al., 2010; Nolan et al., 2008; Vajo, Tamas, Sinka, & Jankovics, 2010). Future studies are therefore needed to investigate this possibility, especially the potential interaction with the influenza vaccine: in the coming years (including the autumn of 2021) COVID-19 vaccination programs will probably overlap with the seasonal Influenza vaccination, so it is crucial to perform additional studies to elucidate the potential interactions and effects of the COVID-19 vaccines with the current vaccination schedules, especially for immunosuppressed and elderly individuals.
The generalizability of these results is subject to certain limitations. First, the number of volunteers in this study was relatively small, although in line with earlier immunological studies on the effects of COVID-19 vaccines. Second, our cohort consisted of healthcare workers, who are middle-aged and healthy, and future studies in elderly individuals and people with comorbidities and other underlying risk factors for severe COVID-19 infections need to be performed (Gao et al., 2021). Third, our study is performed only with individuals with a Western European ancestry. Therefore, the conclusions of our study should be tested in populations with different ancestry and alternative lifestyles since the induction of innate and adaptive immune responses is largely dependent on different factors such as genetic background, diet, and exposure to environmental stimuli which largely differ between communities around the globe.
In conclusion, our data show that the BNT162b2 vaccine induces effects on both the adaptive and the innate branch of immunity and that these effects are different for various SARS-CoV-2 strains. Intriguingly, the BNT162b2 vaccine induces reprogramming of innate immune responses as well, and this needs to be taken into account: in combination with strong adaptive immune responses, this could contribute to a more balanced inflammatory reaction during COVID-19 infection, or it may contribute to a diminished innate immune response towards the virus. BNT162b2 vaccine is clearly protective against COVID-19, but the duration of this protection is not yet known, and one could envisage future generations of the vaccine incorporating this knowledge to improve the range and duration of the protection. Our findings need to be confirmed by conducting larger cohort-studies with populations with diverse backgrounds, while further studies should examine the potential interactions between BNT162b2 and other vaccines.
Author contributions
Conceptualization: M.G.N, F.K.F, J.t.O, J.H., R.v.C, J.v.d.M., F.v.d.V. and L.A.B.J.; Clinical Investigation: F.K.F.; Experimental work: J.D-A, B.G., G.K., O.B., E.S., B.L.H., G.J.O., C.G.v.K., H.D., H.L., S.A.S, M.R.; Supervision: R.P.v.R., M.I.d.J., J.D-A. and M.G.N. Writing and correction of the manuscript: all authors.
Declaration of Interests
M.G.N and L.A.B.J are scientific founders of Trained Therapeutix and Discovery.
Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Mihai G. Netea (mihai.netea{at}radboudumc.nl).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data from this study are available upon request.
Experimental model and subject details
Human subject collection
The study was conducted in compliance with ethical principles of the Declaration of Helsinki, approved by the Arnhem-Nijmegen Institutional Review Board (protocol NL76421.091.21) and registered in the European Clinical Trials Database (2021-000182-33). Health care workers from the Radboudumc Nijmegen were enrolled who received the BNT162b2 mRNA Covid-19 vaccine as per national vaccination campaign and provided informed consent. Subjects (n = 16) were 26-59 years of age, both male and female, and healthy (demographic data presented in Supplementary Table 1). Key exclusion criteria included a medical history of COVID-19. Sera and blood samples were collected before the first administration of BNT162b2, three weeks after the first dose (right before the second dose), and two weeks after the second dose. A high percentage (56.3%) of individuals had been vaccinated with BCG in the past 12 months due to the fact that many participants participated in parallel in a BCG-trial. One individual was removed from the dataset after detecting high concentrations of antibodies against SARS-CoV-2 N-antigen at baseline.
Virus isolation and sequencing
Viruses were isolated from diagnostic specimen at the department of Viroscience, Erasmus MC, and subsequently sequenced to rule out additional mutations in the S protein: D614G (BetaCoV/Munich/BavPat1/2020, European Virus Archive 026V-03883), B.1.1.7 (GISAID: hCov-465 19/Netherlands/ZH-EMC-1148) and B.1.351 (GISAID: hCov-19/Netherlands/ZH-EMC-1461). SARS-CoV-2 isolate BetaCoV/Munich/BavPat1/2020 (European Virus Archive 026V-03883), was kindly provided by Prof. C. Drosten. SARS-CoV-2 Wuhan Hu-1 strain was kindly provided by Prof. Heiner Schaal (Dusseldorf University, Germany). The B.1.1.7 and B.1.351 isolates were isolated from diagnostic specimens on Calu-3 lung adenocarcinoma cells for three passages. Passage 3 BavPat1, B.1.1.7 and B.1.351 variants were used to infect Vero E6 cells at an MOI of 0.01 in DMEM, high glucose (Thermo Fisher Scientific, USA, cat #11965092) supplemented with 2% fetal bovine serum (Sigma-Aldrich, Germany, cat #F7524), 20 mM HEPES (Thermo Fisher Scientific, USA, cat #15630056) and 50 U/mL penicillin-50 µg/mL streptomycin (Thermo Fisher Scientific, USA, cat #15070063) at 37 °C in a humidified 5% CO2 incubator. At 72 h post infection, the culture supernatant was centrifuged for 5 min at 1500 x g and filtered through an 0.45 μM low protein binding filter (Sigma-Aldrich, Germany, cat #SLHPR33RS). To further purify the viral stocks, the medium was transferred over an Amicon Ultra-15 column with 100 kDa cutoff (Sigma-Aldrich, Germany, cat #UFC910008), which was washed 3 times using Opti-MEM supplemented with GlutaMAX (Thermo Fisher Scientific, USA, cat #51985034). Afterwards the concentrated virus on the filter was diluted back to the original volume using Opti-MEM and the purified viral aliquots were stored at -80 °C. The infectious viral titers were measured using plaque assays as described (Varghese et al., 2021) and stocks were heat inactivated for 60 min at 56 °C for use in stimulation experiments.
Measurement of antibody levels against RBD and Spike protein
To measure the levels of antibodies against RBD and Spike protein, a fluorescent-bead-based multiplex immunoassay (MIA) was developed as previously described by (Fröberg et al., 2021). The stabilized pre-fusion conformation of the ectodomain of the Spike protein (amino acids 1 – 1,213) fused with the trimerization motif GCN4 (S-protein) and the receptor binding domain of the S-protein (RBD) were each coupled to beads or microspheres with distinct fluorescence excitation and emission spectra. Serum samples were diluted and incubated with the antigen-coupled microspheres. Following incubation, the microspheres were washed and incubated with phycoerythrin-conjugated goat anti-human, IgG, IgA and IgM. The data were acquired on the Luminex FlexMap3D System. Mean Fluorescent Intensities (MFI) were converted to arbitrary units (AU/ml) by interpolation from a log-5PL-parameter logistic standard curve and log-log axis transformation, using Bioplex Manager 6.2 (Bio-Rad Laboratories) software.
Plaque reduction neutralization assay
A plaque reduction neutralization test (PRNT) was performed. Viruses used in the assay were isolated from diagnostic specimen at the department of Viroscience, Erasmus MC, cultured and subsequently sequenced to rule out additional mutations in the S protein: D614G (GISAID: hCov-19/Netherlands/ZH-EMC-2498), B.1.1.7 (GISAID: hCov-19/Netherlands/ZH-EMC-1148) and B.1.351 (GISAID: hCov-19/Netherlands/ZH-EMC-1461). Heat-inactivated sera were 2-fold diluted in Dulbecco modified Eagle medium supplemented with NaHCO3, HEPES buffer, penicillin, streptomycin, and 1% fetal bovine serum, starting at a dilution of 1:10 in 60 μL. We then added 60 μL of virus suspension (400 plaque-forming units) to each well and incubated at 37°C for 1h. After 1 hour incubation, we transferred the mixtures on to Vero-E6 cells and incubated for 8 hours. After incubation, we fixed the cells with 10% formaldehyde and stained the cells with polyclonal rabbit anti-SARS-CoV antibody (Sino Biological) and a secondary peroxidase-labeled goat anti-rabbit IgG (Dako). We developed signal by using a precipitate forming 3,3′,5,5′-tetramethylbenzidine substrate (True Blue; Kirkegaard and Perry Laboratories) and counted the number of infected cells per well by using an ImmunoSpot Image Analyzer (CTL Europe GmbH). The serum neutralization titer is the reciprocal of the highest dilution resulting in an infection reduction of >50% (PRNT50). We considered a titer >20 to be positive based on assay validation
Isolation of peripheral blood mononuclear cells
Blood samples from subjects were collected into EDTA-coated tubes (BD Bioscience, USA) and used as the source of peripheral blood mononuclear cells (PBMCs) after sampling sera from each individual. Blood is diluted 1:1 with PBS (1X) without Ca++, Mg++ (Westburg, The Netherlands, cat #LO BE17-516F) and PBMCs were isolated via density gradient centrifuge using Ficoll-PaqueTM-plus (VWR, The Netherlands, cat #17-1440-03P). The tubes used for the isolation was specialized SepMate-50 tubes (Stem Cell Technologies, cat #85450) to ensure better separation. Cells counts were determined via Sysmex XN-450 hematology analyzer. Afterwards, PBMCs were frozen using Recovery Cell Culture Freezing (Thermo Fisher Scientific, USA, cat #12648010) in the concentration of 15×106/mL.
Simulation experiments
The PBMCs were thawed and washed with 10mL Dutch modified RPMI 1640 medium (Roswell Park Memorial Institute; Invitrogen, USA, cat # 22409031) containing 50 µg/mL Gentamicine (Centrafarm, The Netherlands), 1 mM Sodium-Pyruvate (Thermo Fisher Scientific, USA, cat #11360088), 2 mM Glutamax (Thermo Fisher Scientific, USA, cat #35050087) supplemented with 10% Bovine Calf Serum (Fisher Scientific, USA, cat #11551831) twice, and afterwards the cells were counted via Sysmex XN-450. PBMCs (4×105 cells/well) stimulated in sterile round bottom 96-well tissue culture treated plates (VWR, The Netherlands, cat #734-2184) in Dutch modified RPMI 1640 medium containing 50 µg/mL Gentamicine, 1 mM Sodium-Pyruvate, 2 mM Glutamax supplemented with 10% human pooled serum. Stimulations were done with heat-inactivated SARS-CoV-2 Wuhan Hu-1 strain (3.3×103 TCID50/mL), SARS-CoV-2 B.1.1.7 (3.3×103 TCID50/mL), SARS-CoV-2 B.1.351 (3.3×103 TCID50/mL), and SARS-CoV-2 Bavarian (3.3×103 TCID50/mL) variants, Influenza (3.3×105 TCID50/mL), 10 µg/mL Poly I:C (Invivogen, USA, cat #tlrl-pic), 3 µg/mL R848 (Invivogen, USA, cat #tlrl-r848), 10 ng/mL E. coli LPS, and 1 x 106 /mL C. albicans. The PBMCs were incubated with the stimulants for 24 hours to detect IL-1β, TNF-α, IL-6, IL-1Ra and 7 days to detect IFN-γ. Supernatants were collected and stored in – 20°C. Secreted cytokine levels from supernatants were quantified by ELISA (IL-1β cat # DLB50, TNF-α cat # STA00D, IL-6 cat # D6050, IL-1Ra cat # DRA00B, IFN-γ cat #DY285B, R&D Systems, USA).
Statistical analysis
Graphpad Prism 8 was used for all statistical analyses. Outcomes between paired groups were analyzed by Wilcoxon’s matched-pairs signed-rank test. Three or more groups were compared using Kruskal-Wallis Test – Dunnet’s multiple comparison. P-value of less than 0.05 was considered statistically significant. Spearman correlation was used to determine correlation between groups.
Acknowledgements
We thank all the volunteers to the study for their willingness to participate. Purified Spike protein and the receptor binding domain of the Spike protein (RBD) were kindly provided by Frank J. van Kuppeveld and Berend-Jan Bosch from the University Utrecht. Y.L. was supported by an ERC Starting Grant (#948207) and the Radboud University Medical Centre Hypatia Grant (2018) for Scientific Research. J.D-A. is supported by The Netherlands Organization for Scientific Research (VENI grant 09150161910024). M.G.N. was supported by an ERC Advanced Grant (#833247) and a Spinoza Grant of the Netherlands Organization for Scientific Research.
Footnotes
-
↵9 Lead contact