As the coronavirus disease 2019 (COVID-19) pandemic continues to take its toll on human life and economic activity, reports have emerged about the repeated positive tests and continued shedding of the virus for weeks and months after clinical recovery.
A study appearing as a preprint on the bioRxiv* server in December 2020 reveals that the genome of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is inserted into the human genome, accounting for the detection of viral RNAs, even in late convalescence.
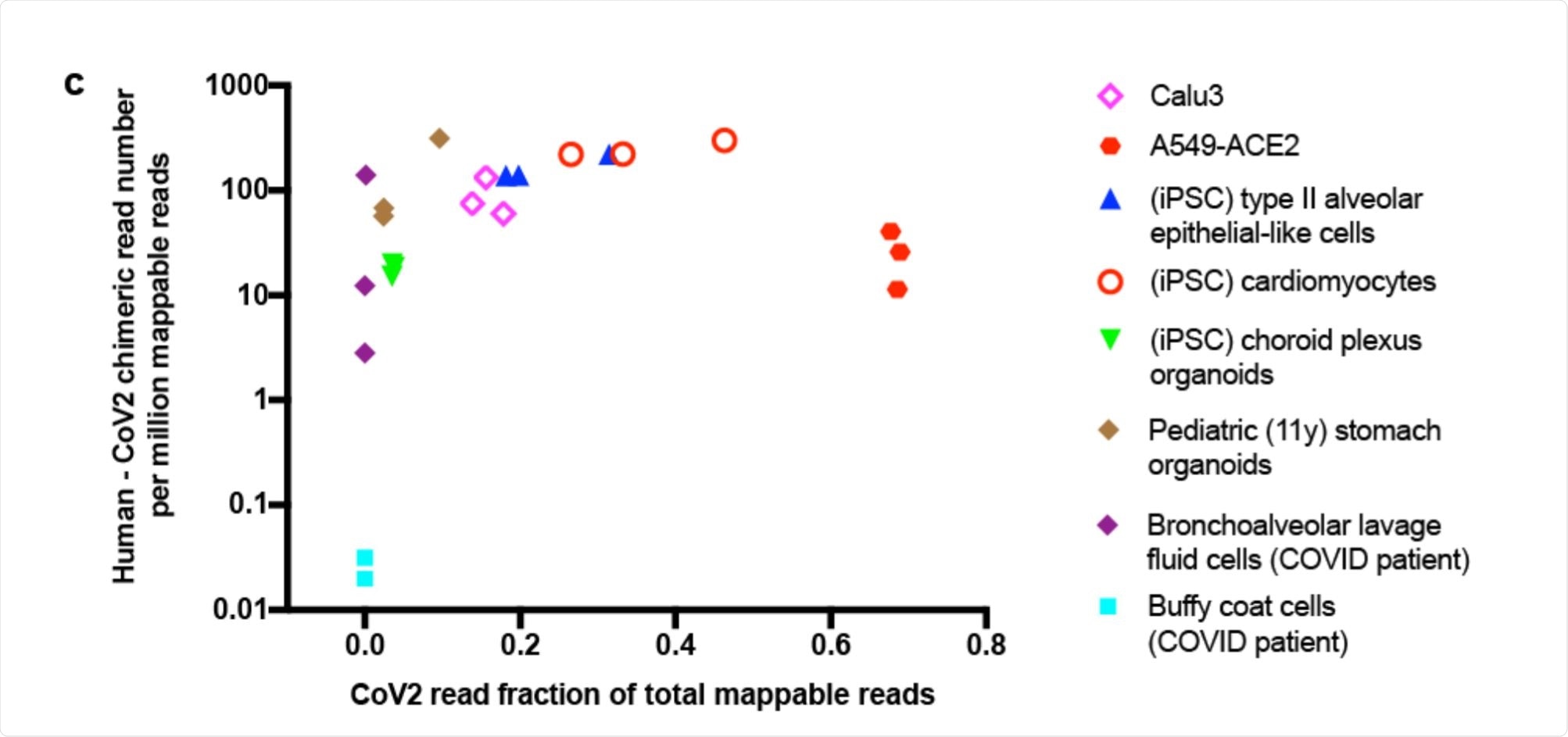
Scatter plot showing human-CoV2 chimeric read number. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.12.422516v1.full.pdf
Reinfection or reverse transcription?
Multiple cases of apparent reinfection have been reported over the past year. Though some have been rigorously investigated and found to be genuine cases of reinfection, evidenced by the presence of different strains of SARS-CoV-2 in the two episodes, it seems unlikely that this is the case with most. Additionally, no replication-competent virus has been isolated.
The current study explored the occurrence of reverse transcription of the SARS-CoV-2 RNA into the human genome. This would result in positive PCR tests due to the continuing transcription of viral RNAs.
Reverse transcriptase (RT) activity has been detected within human cells, as well as integration of the reverse transcription products. For instance, with APP transcripts, their integration into neuronal genomes by endogenous RT was followed by APP transcription.
Such endogenous RT is potentially present in the form of human LINE-1 elements, which make up 17% of the human genome. These are autonomous retrotransposon elements that can transpose themselves as well as other elements of the genome back into the DNA of the nucleus for future transcription.
Chimeric reads present in published RNA sequences
The researchers looked at the published RNA-sequences from SARS-CoV-2 infected cells. Their aim was to find chimeric transcripts, melding human and viral RNA into the same genome. They found a good number of such reads in several different cell types, from the heart, brain, lung and stomach and from cells retrieved from the bronchoalveolar lavage fluid (BALF) obtained from COVID-19 patients.
The proportion of such chimeric sequences was directly correlated with the level of viral RNA in each sample, and such reads typically made up between 0.004% – 0.14% of the total viral reads. The greatest proportion was in BALF cells from severe COVID-19 patients, at ~69%. There were almost none in blood cells, on the other hand.
Most of the host-viral chimeric protein contained the nucleocapsid (N) sequences, as expected since this is the most abundant viral subgenomic RNA. This would, therefore, be the most likely to be reverse transcribed and then integrated. These findings support the occurrence of this event within infected cells.
Sources of RT activity: LINE-1 or HIV-1
The researchers conducted an experiment inducing the overexpression of human LINE-1 elements or HIV-1 RT, in the cell line. The three types of RT examined include: LINE-1 overexpression driven by a CMV promoter, LINE-1 overexpression driven by 5’ UTR, which is its natural promoter, and HIV-1 RT expression.
These cells were then infected with SARS-CoV-2. At two days post-infection, they carried out polymerase chain reaction (PCR) tests to detect the viral sequences, using the N-targeting primer sets used in the commonly used COVID-19 PCR tests.
PCR amplification of the purified cell DNA from infected cells showed the presence of the N protein bands. This did not occur in non-transfected or uninfected cells. Next, they purified the cellular genomic DNA (gDNA) from cells that overexpressed RT and carried out quantitative PCR (qPCR) to confirm the presence of the N sequences.
Overexpression of CMV-LINE-1 led to an 8-fold rise in N-sequence signal strength. This indicated a higher number of N sequences were integrated into the genome in these cells, relative to 5’ UTR-LINE-1 expression or HIV-1 RT expression. They were able to clone the full-length N DNA from cells overexpressing the first RT type, but not the other two. This might be because of the lower number of N sequences integrated into the host genome.
They also conducted an in vitro RT experiment, which showed that cell lysates from cells expressing RT of either type could cause reverse transcription of purified viral RNA from infected cells.
N sequences found in the cell nucleus
Using fluorescent in situ hybridization (FISH) technology, they pinned down the presence and ongoing transcription of the viral N sequences within the cell nucleus with the help of N-targeting fluorescent probes. The N sequences were found in the cytoplasm, as expected of cells infected by SARS-CoV-2.
However, FISH also picked up N RNA signals from the nucleus of cells that overexpressed LINE-1, showing that integrated N sequences in the host genome were being transcribed there.
This occurred in about 35% of cells with overexpressed LINE-1, compared to 12% of non-LINE-1-overexpressing cells. Again, 30% of infected cells that were transfected by LINE-1 plasmids showed FISH nuclear N signals, but only 13% of non-transfected cells. About a tenth of infected non-transfected cells showed nuclear N signals, indicating endogenous RT activity.
LINE-1 and cytokines mediate reverse transcription
The researchers found that published RNA sequencing data from SARS-CoV-2-infected cells showed a high number of LINE-1 elements, which was directly correlated with the abundance of chimeric reads. Within the Calu-3 cell line that allows efficient infection, a number of such elements were upregulated three- to four-fold following infection by the virus. PCR testing showed that these cells demonstrated RT-mediated integration of viral genomic material into the host DNA, perhaps by the activation of the LINE-1 RT. Cytokines can also upregulate endogenous LINE-1 expression two- to three-fold.
Implications and future directions
Our results show induced LINE-1 expression in cells stressed by viral infection or exposed to cytokines, suggesting a molecular mechanism for SARS-CoV-2 retro-integration in human cells.”
The integrated sequences are probably sub-genomic and cannot produce live infectious virions. This explains the positivity of later PCR tests for viral RNA in clinically recovered patients.
More research will be needed to understand whether this will lead to the continuing expression of viral antigens capable of inducing an immune response. It is also possible that the presence of viral elements in the genome could exacerbate the deleterious aspects of the immune response, such as hyper-inflammation, mediated by excessive cytokine release, or autoimmunity.
The authors suggest that the site of insertion and regulation by epigenetic factors, as well as the existing immune state of the patient, may affect the translation of these sequences and their possible clinical consequences. If retro-integration does affect the clinical severity and treatment of COVID-19, the same may possibly be true of other viruses, such as dengue or Zika virus as well, or even the influenza virus.
Finally, the study suggests that many PCR positive results could be due to viral transcripts from such chimeric sequences rather than reflecting the presence of replicating virus in the host. If validated, this will require better tests to be used when assessing the efficacy of COVID-19 therapies in clinical trials, for example, in the future.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Zhang, L. et al. (2020). SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome. bioRxiv preprint. DOI: https://doi.org/10.1101/2020.12.12.422516. https://www.biorxiv.org/content/10.1101/2020.12.12.422516v1
